I am new to both itk/vtk and currently would like to display a 3d render of the vtp file of brain taken converted from nrrd/nifti file, where the final render will be shown in frontend using javascript(react), and the conversation of file from nrrd/nifti could take place in the backend using python.
Hi @babayka,
Its been very frustrated so I want to tell you that to convert an NRRDfile to a VTP file, use Python with the SimpleITK and vtk libraries… First, load the NRRD file with SimpleITK, convert it to a vtkImageData object, then use vtkXMLPolyDataWriter to save it as a VTP file. For frontend rendering, use vtk.js in your React app to load and display the VTP file.
Thanks ![]()
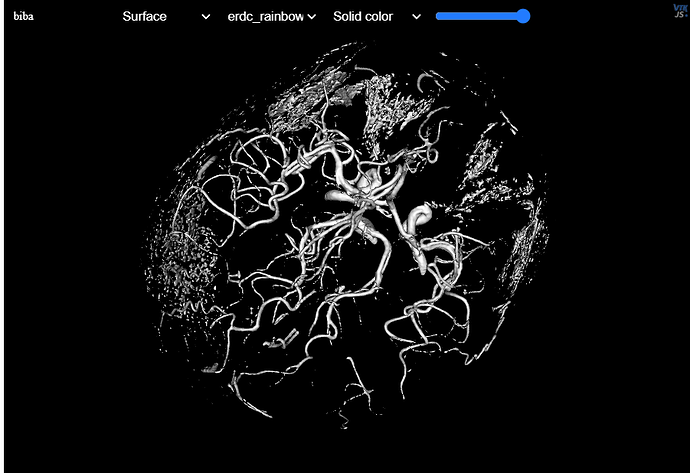
Thanks for the suggestions, I have tried use converting to numpy_array and then into VTKiamgedata from nifti image and then making it into VTP through vtk.vtkMarchingCubes() the issue is that despite it giving, for example, blood vessels through contour_value setting to 150, it missing smaller vessels on the nifti file, is there a way to capture all or at least more vessels without only decreasing the contour_value as with lower values it will just start capturing unnecessary parts of MRA image as well like a
skull, I have attached the image of what I mean
Marching cubes doesn’t handle one or two voxel wide features very well.
You can try vtk’s flying edges and surface nets. Here is a great summary blog https://www.kitware.com/really-fast-isocontouring/
Otherwise you can up sample your images to assign more voxels per feature before extracting the surface.
And using numpy as an intermediary could be bad…it discards voxel spacing and origin info…meaning your extracted vtp data probably won’t align with your original image data in physical space. Itk had built-in converters to and from vtk images, that preserve spacing and origin info…direction info may still be lost / unused by some vtk filters, hopefully not these more modern filters.