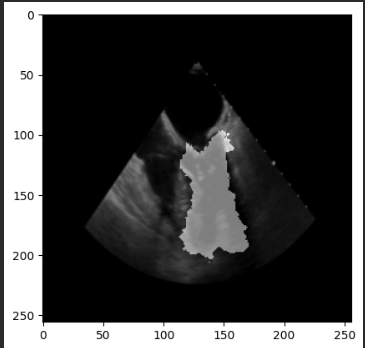
I am wondering how can we create the DisplacementFieldTransform from the output of VoxelMorph. Do we need to change the values/ranges somehow before?
I have this code:
def numpy_to_displacement_field_transform(displacement_field_np):
# Ensure the displacement field is in the correct format: (2, H, W) or (3, H, W, D)
assert len(displacement_field_np.shape) in [3, 4] and displacement_field_np.shape[0] in [2, 3], "Invalid displacement field shape."
# Get the dimension of the displacement field
dimension = displacement_field_np.shape[0]
# Convert the NumPy array to a SimpleITK image
if dimension == 2:
displacement_field_sitk = sitk.GetImageFromArray(np.transpose(displacement_field_np, (1, 2, 0)))
elif dimension == 3:
displacement_field_sitk = sitk.GetImageFromArray(np.transpose(displacement_field_np, (2, 3, 1, 0)))
# Set the pixel type to vector of 64-bit floats
displacement_field_sitk = sitk.Compose(*[sitk.Cast(displacement_field_sitk[:, :, i], sitk.sitkFloat64) for i in range(dimension)])
# Create the DisplacementFieldTransform
displacement_field_transform = sitk.DisplacementFieldTransform(displacement_field_sitk)
return displacement_field_transform
# first segmentation converted as distance map
seg_distance_map = sitk.SignedMaurerDistanceMap(mask_image, squaredDistance=False, useImageSpacing=True)
tx = sitk.Transform(2, sitk.sitkIdentity)
for image_index in range(len(image_paths)):
print(image_index)
next_image = sitk.ReadImage(image_paths[image_index])
moved, displacement_field = model.forward(transform(sitk.GetArrayFromImage(image_with_mask)).to('cuda').unsqueeze(0),
transform(sitk.GetArrayFromImage(next_image)).to('cuda').unsqueeze(0))
latest_displacement_Field_transform = sitk.DisplacementFieldTransform(numpy_to_displacement_field_transform(displacement_field.squeeze(0).squeeze(0).detach().cpu().numpy()))
tx = sitk.DisplacementFieldTransform(sitk.TransformToDisplacementField(sitk.CompositeTransform([tx, latest_displacement_Field_transform]),
size = image_with_mask.GetSize(),
outputOrigin=image_with_mask.GetOrigin(),
outputSpacing = image_with_mask.GetSpacing()))
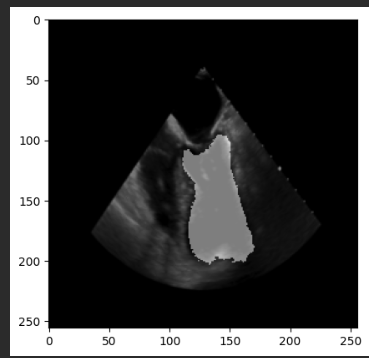
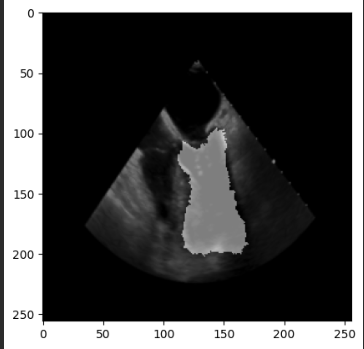
seg_i = sitk.Resample(seg_distance_map, tx) <= 0
seg_image = sitk.GetArrayFromImage(sitk.Cast(seg_i, sitk.sitkUInt8))
plt.imshow(seg_image)
plt.show()
However the output masks are still the same (no changes at all).
Example output from voxelmorph:
tensor([[[[-0.0706, 0.0176, 0.0546, ..., 0.0542, 0.0315, 0.0334],
[-0.0071, 0.0186, 0.0420, ..., 0.0507, 0.0440, 0.0361],
[ 0.0049, -0.0102, -0.0079, ..., 0.0351, 0.0241, 0.0057],
...,
[-0.0484, -0.0361, -0.0431, ..., -0.0435, -0.0262, -0.0348],
[-0.0004, -0.0436, -0.0345, ..., -0.0339, 0.0021, -0.0172],
[-0.0403, -0.0330, -0.0731, ..., -0.0470, -0.0218, -0.0514]],
[[ 0.0106, 0.0601, 0.0193, ..., 0.0139, 0.0019, 0.0044],
[ 0.0086, 0.0362, 0.0331, ..., 0.0609, 0.0225, 0.0146],
[ 0.0224, 0.0382, 0.0524, ..., 0.0538, 0.0156, 0.0011],
...,
[ 0.0170, 0.0226, 0.0176, ..., -0.0324, -0.0620, -0.0514],
[-0.0039, -0.0075, -0.0010, ..., -0.0253, -0.0645, -0.0503],
[-0.0311, 0.0211, -0.0075, ..., -0.0388, -0.0689, -0.0073]]]],
device='cuda:0', grad_fn=<AliasBackward0>)
max displacement: tensor(9.1939, device='cuda:0', grad_fn=<AliasBackward0>)
min displacement: tensor(-10.1772, device='cuda:0', grad_fn=<AliasBackward0>)
example npy def. field attached:
test.npy (512.1 KB)
 ]
]