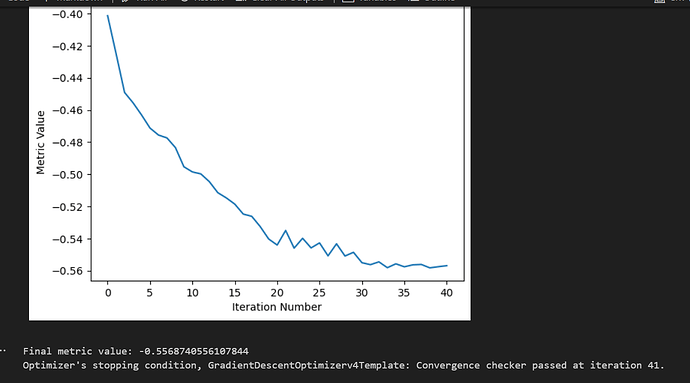
Hi, I want to perform affine registration on 3D heart CT images obtained from different individuals. Firstly, I resampled all images to a uniform size of 111 and then cropped them to ensure the heart is centered and this makes images have different sizes. Subsequently, the images were normalized to have intensities ranging from 0 to 2048. After all, I performed affine registration, but the result is not satisfactory and I am wondering how I can improve it?
type oriteration_number = []
metric_values = []
def update_metric_plot():
iteration_number.append(registration_method.GetOptimizerIteration())
metric_values.append(registration_method.GetMetricValue())
add_fix = "/home/sargordis/Desktop/convert/resample111/best/crop/fix/0.nii.gz"
add_moving = "/home/sargordis/Desktop/convert/resample111/best/crop/moving/1.nii.gz"
fixed_image = sitk.ReadImage(add_fix, sitk.sitkFloat32)
moving_image = sitk.ReadImage(add_moving, sitk.sitkFloat32)
initial_transform = sitk.CenteredTransformInitializer(fixed_image,
moving_image,
sitk.AffineTransform(3),
sitk.CenteredTransformInitializerFilter.MOMENTS)
#sitk.CenteredTransformInitializerFilter.GEOMETRY)
resampled_moving = sitk.Resample(moving_image, fixed_image, initial_transform)
registration_method = sitk.ImageRegistrationMethod()
#registration_method.SetMetricAsCorrelation()
registration_method.SetMetricAsMattesMutualInformation(numberOfHistogramBins=50)
registration_method.SetMetricSamplingStrategy(registration_method.RANDOM)
registration_method.SetMetricSamplingPercentage(0.01)
registration_method.SetInterpolator(sitk.sitkLinear)
registration_method.SetOptimizerAsGradientDescent(learningRate=0.001, numberOfIterations=300, convergenceMinimumValue=1e-6, convergenceWindowSize=10)
registration_method.SetOptimizerScalesFromPhysicalShift()
# # Setup for the multi-resolution framework.
# registration_method.SetShrinkFactorsPerLevel(shrinkFactors=[4, 2, 1])
# registration_method.SetSmoothingSigmasPerLevel(smoothingSigmas=[2, 1, 0])
# registration_method.SmoothingSigmasAreSpecifiedInPhysicalUnitsOn()
registration_method.SetInitialTransform(initial_transform, inPlace=False)
registration_method.AddCommand(sitk.sitkIterationEvent, lambda: update_metric_plot())
final_transform = registration_method.Execute(fixed_image , moving_image)
print(registration_method.GetOptimizerStopConditionDescription())
out = sitk.Resample(moving_image, fixed_image, final_transform, sitk.sitkLinear, 0.0, moving_image.GetPixelID())
#moved_image = sitk.Resample(moving_image, fixed_image, final_transform, sitk.sitkLinear, 0.0)
sitk.WriteImage(out, '/home/sargordis/Desktop/moved.nii.gz')
plt.plot(iteration_number, metric_values)
plt.xlabel('Iteration Number')
plt.ylabel('Metric Value')
plt.title('Metric Evolution During Registration')
plt.show()
# Always check the reason optimization terminated.
print("Final metric value: {0}".format(metric_values[-1]))
print(
"Optimizer's stopping condition, {0}".format(
registration_method.GetOptimizerStopConditionDescription()
)
)
paste code here
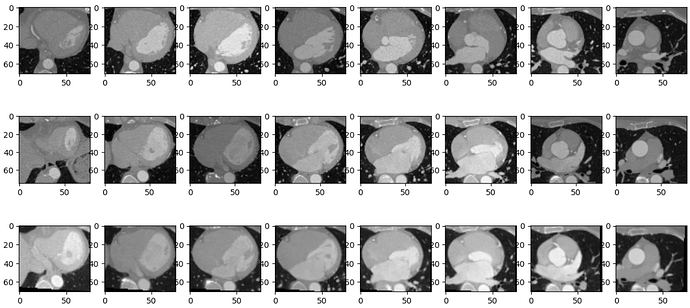
In the below image, the first row is fixed, the middle is moving and 3rd row is the moved image.