Hello Everyone,
I have done registration between MRI and CT scans, and I wanna save the output of registered slices?
import matplotlib.pyplot as plt
import SimpleITK as sitk
from ipywidgets import interact, fixed
from IPython.display import clear_output
from myshow import myshow
import numpy as np
import gui
import sys
from downloaddata import fetch_data as fdata
import registration_gui as rgui
import os
OUTPUT_DIR = 'output'
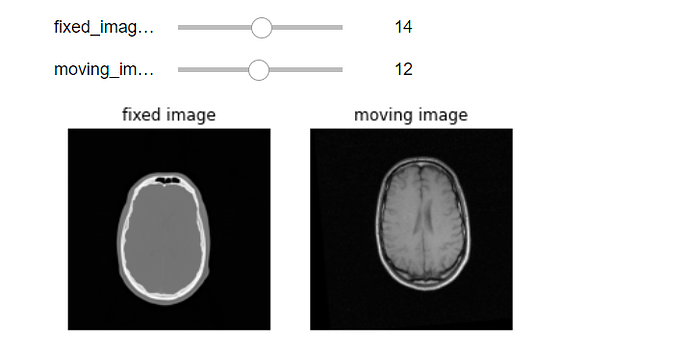
def display_images(fixed_image_z, moving_image_z, fixed_npa, moving_npa):
# Create a figure with two subplots and the specified size.
#plt.subplots(1, 2, figsize=(10, 8))
# Draw the fixed image in the first subplot.
plt.subplot(1, 2, 1)
plt.imshow(fixed_npa[fixed_image_z, :, :], cmap=plt.cm.Greys_r);
plt.title('fixed image')
plt.axis('off')
# Draw the moving image in the second subplot.
plt.subplot(1, 2, 2)
plt.imshow(moving_npa[moving_image_z, :, :], cmap=plt.cm.Greys_r);
plt.title('moving image')
plt.axis('off')
plt.show()
# Callback invoked by the IPython interact method for scrolling and modifying the alpha blending
# of an image stack of two images that occupy the same physical space.
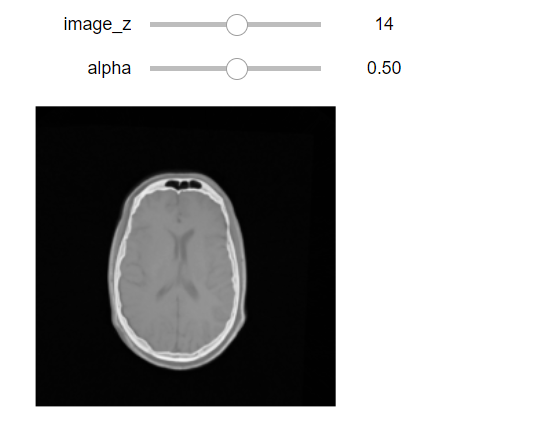
def display_images_with_alpha(image_z, alpha, fixed, moving):
img = (1.0 - alpha) * fixed[:, :, image_z] + alpha * moving[:, :, image_z]
plt.imshow(sitk.GetArrayViewFromImage(img), cmap=plt.cm.Greys_r);
plt.axis('off')
plt.show()
# Callback invoked when the StartEvent happens, sets up our new data.
def start_plot():
global metric_values, multires_iterations
metric_values = []
multires_iterations = []
# Callback invoked when the EndEvent happens, do cleanup of data and figure.
def end_plot():
global metric_values, multires_iterations
del metric_values
del multires_iterations
# Close figure, we don't want to get a duplicate of the plot latter on.
plt.close()
# Callback invoked when the IterationEvent happens, update our data and display new figure.
def plot_values(registration_method):
global metric_values, multires_iterations
metric_values.append(registration_method.GetMetricValue())
# Clear the output area (wait=True, to reduce flickering), and plot current data
clear_output(wait=True)
# Plot the similarity metric values
plt.plot(metric_values, 'r')
plt.plot(multires_iterations, [metric_values[index] for index in multires_iterations], 'b*')
plt.xlabel('Iteration Number', fontsize=12)
plt.ylabel('Metric Value', fontsize=12)
plt.show()
# Callback invoked when the sitkMultiResolutionIterationEvent happens, update the index into the
# metric_values list.
def update_multires_iterations():
global metric_values, multires_iterations
multires_iterations.append(len(metric_values))
####################################Their own data ################################################
#fixed_image = sitk.ReadImage(fdata("training_001_ct.mha"), sitk.sitkFloat32)
#moving_image = sitk.ReadImage(fdata("training_001_mr_T1.mha"), sitk.sitkFloat32)
####################################Their own data ################################################
########################################### Hospital data #################################
data_directory = r"CT_1"
series_IDs = sitk.ImageSeriesReader.GetGDCMSeriesIDs(data_directory)
if not series_IDs:
print("ERROR: given directory \"" + data_directory + "\" does not contain a DICOM series.")
sys.exit(1)
series_file_names = sitk.ImageSeriesReader.GetGDCMSeriesFileNames(data_directory, series_IDs[0])
data_directory2 = r"MR_1"
series_IDs2 = sitk.ImageSeriesReader.GetGDCMSeriesIDs(data_directory2)
if not series_IDs2:
print("ERROR: given directory \"" + data_directory2 + "\" does not contain a DICOM series.")
sys.exit(1)
series_file_names2 = sitk.ImageSeriesReader.GetGDCMSeriesFileNames(data_directory2, series_IDs2[0])
fixed_image = sitk.ReadImage(series_file_names, sitk.sitkFloat32)
moving_image = sitk.ReadImage(series_file_names2, sitk.sitkFloat32)
########################################### Hospital data #################################
initial_transform = sitk.CenteredTransformInitializer(fixed_image,
moving_image,
sitk.ScaleVersor3DTransform(),
sitk.CenteredTransformInitializerFilter.GEOMETRY)
moving_resampled = sitk.Resample(moving_image, fixed_image, initial_transform, sitk.sitkLinear, 0.0, moving_image.GetPixelID())
registration_method = sitk.ImageRegistrationMethod()
# Similarity metric settings.
registration_method.SetMetricAsMattesMutualInformation(numberOfHistogramBins=50)
registration_method.SetMetricSamplingStrategy(registration_method.RANDOM)
registration_method.SetMetricSamplingPercentage(0.01)
registration_method.SetInterpolator(sitk.sitkLinear)
# Optimizer settings.
registration_method.SetOptimizerAsGradientDescent(learningRate=1.0, numberOfIterations=100, convergenceMinimumValue=1e-6,
convergenceWindowSize=10)
registration_method.SetOptimizerScalesFromPhysicalShift()
# Setup for the multi-resolution framework.
#registration_method.SetShrinkFactorsPerLevel(shrinkFactors = [4,2,1])
#registration_method.SetSmoothingSigmasPerLevel(smoothingSigmas=[2,1,0])
#registration_method.SmoothingSigmasAreSpecifiedInPhysicalUnitsOn()
# Don't optimize in-place, we would possibly like to run this cell multiple times.
registration_method.SetInitialTransform(initial_transform, inPlace=False)
# Connect all of the observers so that we can perform plotting during registration.
registration_method.AddCommand(sitk.sitkStartEvent, start_plot)
registration_method.AddCommand(sitk.sitkEndEvent, end_plot)
registration_method.AddCommand(sitk.sitkMultiResolutionIterationEvent, update_multires_iterations)
registration_method.AddCommand(sitk.sitkIterationEvent, lambda: plot_values(registration_method))
final_transform = registration_method.Execute(sitk.Cast(fixed_image, sitk.sitkFloat32),
sitk.Cast(moving_image, sitk.sitkFloat32))
print(f'Final metric value: {registration_method.GetMetricValue()}')
print(f'Optimizer\'s stopping condition, {registration_method.GetOptimizerStopConditionDescription()}')
moving_resampled = sitk.Resample(moving_image, fixed_image, final_transform, sitk.sitkLinear, 0.0, moving_image.GetPixelID())
interact(display_images, fixed_image_z=(0,fixed_image.GetSize()[2]-1), moving_image_z=(0,moving_image.GetSize()[2]-1),
fixed_npa = fixed(sitk.GetArrayViewFromImage(fixed_image)), moving_npa=fixed(sitk.GetArrayViewFromImage(moving_image)));
interact(display_images_with_alpha, image_z=(0,fixed_image.GetSize()[2] - 1), alpha=(0.0,1.0,0.05),
fixed = fixed(fixed_image), moving=fixed(moving_resampled));
sitk.WriteImage(moving_resampled, os.path.join(OUTPUT_DIR, 'RIRE_training_001_mr_T1_resampled.mha'))
#print(moving_resampled)
sitk.WriteTransform(final_transform, os.path.join(OUTPUT_DIR, 'RIRE_training_001_CT_2_mr_T1.tfm'))
Inputs:
Output: