Hi @zivy,
I have been using the BSplineSTN3D model from here which is based in BSplineTransformation from here. The NN outputs the parameters and those parameters are used to deform the image. I applied those parameters in BSplineTransform of SITK which results in a different image. The mentioned code has methods to compute the grid using the displacement field. They are using some convolutional operations and ultimately reduce to the shape of the volume that is being used.
I also wanted to visualize the deformation field thinking that might help. Is there some tool in SITK to visualize grid?
import SimpleITK as sitk
import torch
import numpy as np
import pickle
with open("bspline_params.pkl", "rb") as f:
bspline_params = pickle.load(f).detach().cpu().numpy().squeeze()
img = sitk.ReadImage("mri.nii")
image_size = img.GetSize()
spacing = (10, 10, 10)
mesh_size = np.ceil(np.array(image_size) / spacing).astype(np.int).tolist()
bspline_transform = sitk.BSplineTransform(3, 2)
bspline_transform.SetTransformDomainPhysicalDimensions(img.GetSize())
bspline_transform.SetTransformDomainMeshSize(mesh_size)
bspline_transform.SetTransformDomainDirection(img.GetDirection())
bspline_transform.SetTransformDomainOrigin(img.GetOrigin())
bspline_params_list = bspline_params.flatten().tolist()
bspline_transform.SetParameters(bspline_params_list)
resampled_image = sitk.Resample(img, img, bspline_transform, sitk.sitkNearestNeighbor, 0.0)
sitk.WriteImage(resampled_image, "transformed_using_sitk.nii")
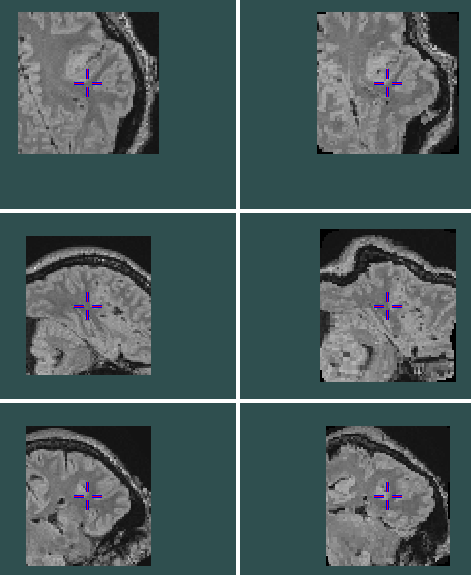
The output is attached below(left is sitk transformation and is close to original image, right is Pytorch transformation which seems heavily deformed)
this is the parameters dump used and this is the image
I had posted another question in the project repo. Turns out, we cannot relate these transformations. Now I am wondering whether I am using the BSplineTransform correctly or not since it does not seem to warp the image.