Hi all,
I have a question regarding masking in non-rigid registration. So I have already done a rigid registration of two histological slices, that are now globally well-disposed (at the first glance). However, due to the level of detail and size of these histo images, local structures within the tissue are not as good aligned as the whole particle. When I run a non-rigid deformation for that purpose now, I face the problem that the tissue might deform excessively what leads to an information loss consequntly. Trying to penalize the deformation didn’t solve my problem completely.
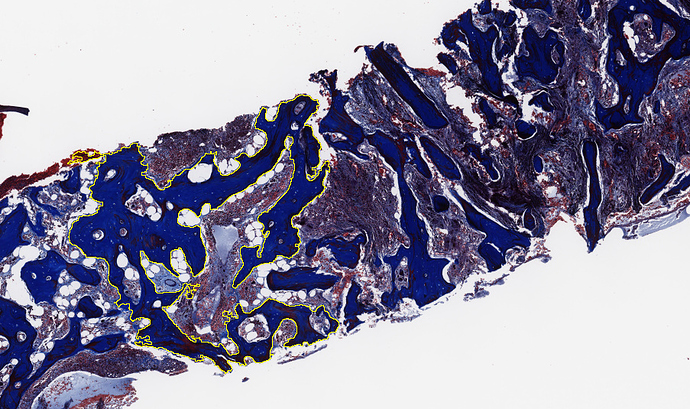
So I was wondering if I could use a property of my spcific tissue for the registration. The image data are bone marrow slices with a lot of bone structures inside the tissue (see image; yellow area). Now my idea is to make a pre-segmentaion of the bone structures inside the particle and a second segmentation of the marrow inbetween. Then run a combined registration for both of these masked areas, allowing the bone structures to deform as much as they like while penalizing/keeping the deformation energy of the marrow small. In brief: I want to keep the local deformation of the marrow as less as possible but do all relevant transformations to get a satisfying registration result.
(The yellow area shows one of those typical bone structures that have no further significance for analysis. The marrow tissue right of it / inbetween should only be transformed as much as necessary.)
I hope that i have been able to clearify my concern and am looking forward to your suggestions and ideas.
Thanks in advance and best regards,
Michael