Hello all,
I am using ITK to resample CT 3D volume. I have done two version, one is c++, one is python. The difference between their result is not small. The bigger difference is existed in boundary part in the image.BTW, both of them used linear interpolator.
As I know, the python itk is wrapped on the c++ library. Am I right? So they should not have difference from in theory. I have also tried the SimpleITK in python. There is no difference between simpleITK and ITK results in python.
Here is code segments :
def resize_itk(data, new_shape, order=3):
# Resample images to 2mm spacing with ITK
import itk
itk_image=itk.GetImageFromArray(data)
itk_image.SetSpacing( [0.791,0.791,1] )
original_spacing = itk.spacing(itk_image)
original_size =itk.size(itk_image)
dimension = itk_image.GetImageDimension()
out_spacing = [2.0,2.0,2.0]
# out_size = [203,203,396]
out_size = [
int(np.round(original_size[dim] * (original_spacing[dim] / out_spacing[dim])+0.5)) for dim in range(dimension)
]
transform = itk.IdentityTransform[itk.D, dimension].New()
ImageType = type(itk_image)
interpolator = itk.LinearInterpolateImageFunction[ImageType, itk.D].New()
out_image = itk.resample_image_filter(
itk_image,
# interpolator = interpolator,
size=out_size,
output_spacing=out_spacing,
output_origin=itk.origin(itk_image),
transform=transform,
)
itk.imwrite(out_image, "/mnt/store/data/BR_DATASET/resampled-839316-py-itk-default-interpolator-itkF.nii.gz")
return itk.GetArrayFromImage(out_image)
def resize_sitk(data, new_shape, order=3):
# Resample images to 2mm spacing with SimpleITK
import SimpleITK as sitk
itk_image = sitk.GetImageFromArray(data)
itk_image.SetSpacing( [0.791,0.791,1] )
original_spacing = itk_image.GetSpacing()
original_size = itk_image.GetSize()
out_spacing = [2.0,2.0,2.0]
# out_size = [203,203,396]
out_size = [
int(np.round(original_size[0] * (original_spacing[0] / out_spacing[0])+0.5)),
int(np.round(original_size[1] * (original_spacing[1] / out_spacing[1])+0.5)),
int(np.round(original_size[2] * (original_spacing[2] / out_spacing[2])+0.5))]
resample = sitk.ResampleImageFilter()
resample.SetOutputSpacing(out_spacing)
resample.SetSize(out_size)
resample.SetOutputDirection(itk_image.GetDirection())
resample.SetOutputOrigin(itk_image.GetOrigin())
resample.SetTransform(sitk.Transform())
resample.SetDefaultPixelValue(itk_image.GetPixelIDValue())
resample.SetInterpolator(sitk.sitkLinear)
out_img = resample.Execute(itk_image)
sitk.WriteImage(out_img,"/mnt/store/data/BR_DATASET/resampled-839316-py-sitk.nii.gz")
return sitk.GetArrayFromImage(out_img)
template< typename InputPixelType, typename OutputPixelType, typename InterpolatorPrecisionType = double,
typename TransformPrecisionType = InterpolatorPrecisionType, uint16_t dim = 3 >
static int Resampling(itk::SmartPointer<itk::Image<InputPixelType, dim>>& InputImg,
double vfOutputSpacing[], const double vfOutputOrigin[],itk::Size<dim>& vnOutputSize,
itk::SmartPointer<itk::Image<OutputPixelType, dim>>& OutputImg,uint8_t nInterpolator=0)
{
using InputImageType = itk::Image<InputPixelType, dim>;
using OutputImageType = itk::Image<OutputPixelType, dim>;
//auto inputRegion = InputImg->GetLargestPossibleRegion();
//auto vnInputSize = inputRegion.GetSize();
//auto oriSpacing = InputImg->GetSpacing();
//-----Resampling------------------------------------------------
// Identity transform
using T_Transform = itk::IdentityTransform<TransformPrecisionType, dim>;
// The resampler type itself.
using T_ResampleFilter = itk::ResampleImageFilter<InputImageType, OutputImageType,
InterpolatorPrecisionType, TransformPrecisionType>;
// Prepare the resampler.
// Instantiate the transform and specify it should be the id transform.
T_Transform::Pointer pTransform = T_Transform::New();
pTransform->SetIdentity();
// Instantiate the resampler. Wire in the transform and the interpolator.
T_ResampleFilter::Pointer pResizeFilter = T_ResampleFilter::New();
pResizeFilter->SetTransform(pTransform);
// If ITK resampler determines there is something to interpolate which is
// usually the case when upscaling (!) then we must specify the interpolation algorithm.
if (nInterpolator == LINEAR)
{
using T_Interpolator = itk::LinearInterpolateImageFunction<InputImageType, InterpolatorPrecisionType>;
// Instantiate the linear interpolator.
T_Interpolator::Pointer pInterpolator = T_Interpolator::New();
pResizeFilter->SetInterpolator(pInterpolator);
}
else if (nInterpolator == BSPLINE)
{
//default order is 3
using T_Interpolator = itk::BSplineInterpolateImageFunction<InputImageType, InterpolatorPrecisionType,
InterpolatorPrecisionType>;
T_Interpolator::Pointer pInterpolator = T_Interpolator::New();
//pInterpolator->SetSplineOrder(3);
pResizeFilter->SetInterpolator(pInterpolator);
}
else
{
mippVolumeEngineLog::GetInstance()->PrintDebugInfo(LogInfoType::Information,
L"The interplator is unexpected.", std::to_wstring(UnexpInterplatorErr));
return UnexpInterplatorErr;
}
//if (nInterpolator == NEAREST)
//{
// using T_Interpolator = itk::NearestNeighborInterpolateImageFunction<InputImageType, double>;
// T_Interpolator::Pointer pInterpolator = T_Interpolator::New();
// pResizeFilter->SetInterpolator(pInterpolator);
//}
// Set the output origin. You may shift the original image "inside" the
// new image size by specifying something else than 0.0, 0.0 here.
pResizeFilter->SetOutputOrigin(vfOutputOrigin);
// Set the output spacing.
pResizeFilter->SetOutputSpacing(vfOutputSpacing);
// Set the output size.
pResizeFilter->SetSize(vnOutputSize);
pResizeFilter->SetInput(InputImg);
pResizeFilter->Update();
OutputImg = pResizeFilter->GetOutput();
return Success;
}
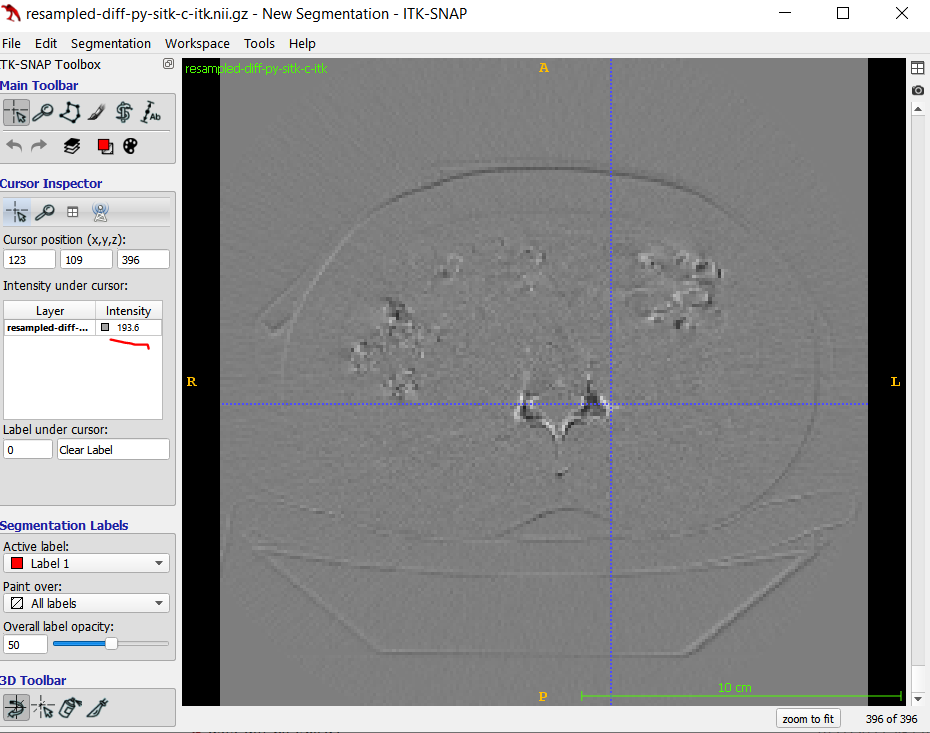
Here is one slice in the difference result:
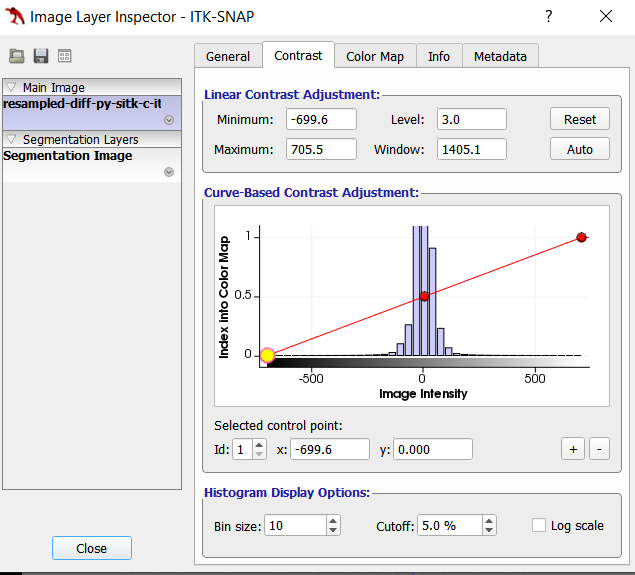
Here is the histogram of the difference result:
Is there any problems in the code? Can I get the same result in python with c++?
Looking forward to get your reply. Thanks a lot.