Hello I am working with dicom and nrrd image file formats. Usually nrrd files are segmentations and in different spacing and origin compared to the dicoms. So my workflow is:
- resample the nrrd file on dicom to match dicom’s spacing and origin and other image properties.
- Then perform some morphological operation on the resampled nrrd file.
- convert both dicom and resampled nrrd to nifti format maintaining the properties.
For these 3 steps I am using the following:
reader = sitk.ImageSeriesReader()
dicom_names = reader.GetGDCMSeriesFileNames(dcmCasePath)
reader.SetFileNames(dicom_names)
dicom = reader.Execute()
nrrd = sitk.ReadImage(nrrdFilePath)
use the following function to resample nrrd on dicom:
def Realign(self, image, reference_image, def_value=0.0):
"""
:param image: image to resample or align(.nrrd files)
:param reference_image: .dcm files
:param def_value: values to fill the resampled image
:return: aligned image that matches reference image origin, spacing etc.
"""
return sitk.Resample(image,
reference_image,
sitk.Transform(),
sitk.sitkLinear,
def_value,
reference_image.GetPixelID())
data = [dicom, nrrd]
alignedNrd= Realign(data[1], data[0])
Then I perform some morphological operations on NumPy-based aligned image.
SegBN = np.zeros_like(sitk.GetArrayFromImage(alignedNrd))
SegBN[np.where(sitk.GetArrayFromImage(alignedNrd) > 0)] = 1
Finally saving the image in nifti:
nrd2niiPath = os.path.splitext(nrdpath)[0]+'_{}'.format(ig)+'.nii.gz'
sitk.WriteImage(sitk.GetImageFromArray(SegBN), nrd2niiPath)
dcm2niiPath = os.path.join(dcmPath, os.path.basename(dcmPath)) +'.nii.gz'
sitk.WriteImage(dicom, dcm2niiPath)
But the spacing and origin in the nifti images are totally different. Despite the nrrd image was resampled in reference to the dicom.
To check:
dcmimage.GetSpacing(), alignedNrrd.GetSpacing()
((0.3613280058, 0.3613280058, 0.625), (0.3613280058, 0.3613280058, 0.625))
dcmimage.GetOrigin(), alignedNrrd.GetOrigin()
((-69.19999695, -109.8000031, -240.375),
(-69.19999695, -109.8000031, -240.375))
dcmimage.GetSize(), alignedNrrd.GetSize()
((512, 512, 224), (512, 512, 224))
imgnb.get_affine(), segnb.get_affine()
(array([[ -0.36132801, 0. , 0. , 69.19999695],
[ 0. , -0.36132801, 0. , 109.80000305],
[ 0. , 0. , 0.625 , -240.375 ],
[ 0. , 0. , 0. , 1. ]]),
array([[-1., 0., 0., -0.],
[ 0., -1., 0., -0.],
[ 0., 0., 1., 0.],
[ 0., 0., 0., 1.]]))
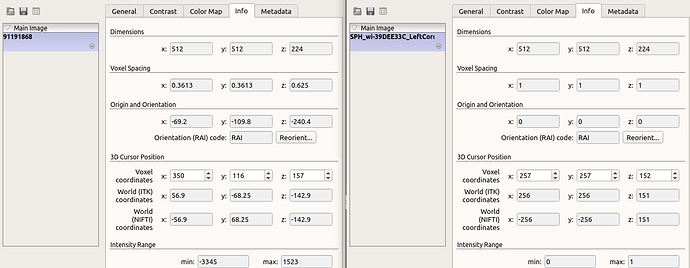
As it can be seen the nifti formats are in different spacings.
To visualize the nifti image and segmentation separately in ITKsnap:
For some reason, the segmentation is isotropic and completely in different spacing and origin, despite the image size are the same.
What is happening after the morphological operation or saving niftis that changes the image origin, spacing and all?