Hello everyone,
I’m trying to compute the Dice coefficient using a Python script.
To implement this, I have used ideas and code from the following links:
“SlicerRT/SegmentComparison at master · SlicerRt/SlicerRT · GitHub”
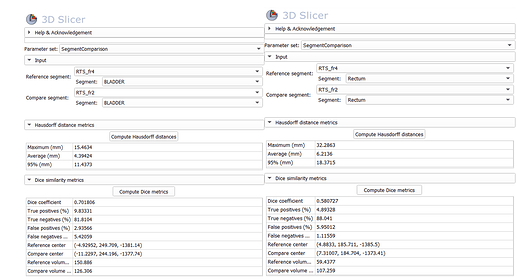
However, as shown in the attached screenshot, there is a slight difference between the Dice coefficient values computed by my code and those reported by 3D Slicer
(my results:
Dice for Rectum=0.628, Bladder= 0.749)
Could you please help me understand why there is this discrepancy and how I can resolve it so that my results match Slicer’s output?
Thanks a lot
my code:
import SimpleITK as sitk
import numpy as np
import pydicom
import logging
import os
from skimage.draw import polygon
from scipy.spatial.distance import directed_hausdorff
from scipy.ndimage import binary_dilation
logging.basicConfig(level=logging.INFO, format=‘%(asctime)s - %(levelname)s - %(message)s’)
def read_dicom_series(directory):
if not os.path.isdir(directory): raise FileNotFoundError(f"Directory not found: {directory}“)
series_IDs = sitk.ImageSeriesReader.GetGDCMSeriesIDs(directory)
if not series_IDs: raise ValueError(f"No DICOM series found in: {directory}”)
series_files = sitk.ImageSeriesReader.GetGDCMSeriesFileNames(directory, series_IDs[0])
reader = sitk.ImageSeriesReader(); reader.SetFileNames(series_files)
return reader.Execute()
def list_rois(rtstruct_path):
ds = pydicom.dcmread(rtstruct_path)
return [roi.ROIName for roi in ds.StructureSetROISequence] if hasattr(ds, ‘StructureSetROISequence’) else [ ]
def extract_mask_from_rtstruct(rtstruct_path, roi_name, reference_ct, output_nifti=None):
ds = pydicom.dcmread(rtstruct_path)
roi_number = next((i+1 for i, roi in enumerate(ds.StructureSetROISequence) if roi.ROIName.lower() == roi_name.lower()), None)
if not roi_number:
logging.warning(f"ROI ‘{roi_name}’ not found in {rtstruct_path}.“)
return None
contour_seq = next((rc.ContourSequence for rc in ds.ROIContourSequence if rc.ReferencedROINumber == roi_number), None)
if not contour_seq:
logging.warning(f"No contours found for ‘{roi_name}’ in {rtstruct_path}.”)
return None
mask_array = np.zeros(reference_ct.GetSize()[::-1], dtype=np.uint8) # z, y, x
has_contours = False
for contour in contour_seq:
points = np.array(contour.ContourData).reshape(-1, 3)
if len(points) < 3: continue
has_contours = True
z_pos = points[0, 2]
slice_index = int(round(reference_ct.TransformPhysicalPointToContinuousIndex((0, 0, z_pos))[2]))
if not (0 <= slice_index < mask_array.shape[0]):
logging.warning(f"Slice index {slice_index} out of bounds - skipping.“)
continue
rows, cols = [ ], [ ]
for p in points:
cont_idx = reference_ct.TransformPhysicalPointToContinuousIndex(p)
rows.append(cont_idx[1])
cols.append(cont_idx[0])
rr, cc = polygon(rows, cols, shape=mask_array.shape[1:])
mask_array[slice_index, rr, cc] = 1
if not has_contours:
logging.warning(f"No valid contours for ‘{roi_name}’ - returning None.”)
return None
####
mask_array = binary_dilation(mask_array, structure=np.ones((3,3,3)))
mask = sitk.GetImageFromArray(mask_array.astype(np.uint8))
mask = sitk.BinaryFillhole(mask) # Fill holes like Slicer
mask.CopyInformation(reference_ct)
if output_nifti:
sitk.WriteImage(mask, output_nifti)
logging.info(f"Mask saved to: {output_nifti}“)
return mask
def compute_dice(fixed_mask, moving_mask):
overlap_filter = sitk.LabelOverlapMeasuresImageFilter()
overlap_filter.Execute(fixed_mask, moving_mask)
return overlap_filter.GetDiceCoefficient()
def compute_hausdorff_distance(fixed_mask, moving_mask):
fixed_points = np.argwhere(sitk.GetArrayFromImage(fixed_mask) > 0)
moving_points = np.argwhere(sitk.GetArrayFromImage(moving_mask) > 0)
if fixed_points.size == 0 or moving_points.size == 0:
return float(‘inf’)
dist1 = directed_hausdorff(fixed_points, moving_points)[0]
dist2 = directed_hausdorff(moving_points, fixed_points)[0]
return max(dist1, dist2)
def compute_overlap_fraction(fixed_mask, moving_mask):
intersection = np.logical_and(sitk.GetArrayFromImage(fixed_mask), sitk.GetArrayFromImage(moving_mask)).sum()
return intersection / (sitk.GetArrayFromImage(fixed_mask).sum() + 1e-6)
def main():
fixed_ct_dir = r’D:\fr4\CT_Unified’
fixed_rtstruct_path = r’D:\fr4\STRUCT_Unified\RTS_fr4.dcm’
transformed_rtstruct_path = r’D:\fr2\STRUCT_Unified\RTS_fr2.dcm’
fixed_ct = read_dicom_series(fixed_ct_dir)
fixed_rois = list_rois(fixed_rtstruct_path)
transformed_rois = list_rois(transformed_rtstruct_path)
logging.info(”\nAvailable ROIs in fixed RTSTRUCT:“)
for roi in fixed_rois: logging.info(f” - {roi}“)
logging.info(”\nAvailable ROIs in transformed RTSTRUCT:“)
for roi in transformed_rois: logging.info(f” - {roi}“)
organs = [‘Bladder’, ‘Rectum’]
for organ in organs:
try:
fixed_roi = input(f"Enter ROI name for {organ} in fixed RTSTRUCT (from list above): “).strip()
transformed_roi = input(f"Enter ROI name for {organ} in transformed RTSTRUCT (from list above): “).strip()
fixed_mask = extract_mask_from_rtstruct(fixed_rtstruct_path, fixed_roi, fixed_ct, f"fixed_{organ}mask.nii.gz")
transformed_mask = extract_mask_from_rtstruct(transformed_rtstruct_path, transformed_roi, fixed_ct, f"transformed{organ}_mask.nii.gz”)
if fixed_mask is None or transformed_mask is None:
logging.warning(f"Skipping metrics for ‘{organ}’ due to missing mask.”)
continue
resampler = sitk.ResampleImageFilter()
resampler.SetReferenceImage(fixed_mask)
resampler.SetInterpolator(sitk.sitkNearestNeighbor)
transformed_mask = resampler.Execute(transformed_mask)
dice = compute_dice(fixed_mask, transformed_mask)
hausdorff = compute_hausdorff_distance(fixed_mask, transformed_mask)
overlap = compute_overlap_fraction(fixed_mask, transformed_mask)
logging.info(f”\nMetrics for {organ} (fixed: ‘{fixed_roi}’, transformed: ‘{transformed_roi}’):“)
logging.info(f” Dice Coefficient: {dice:.4f}“)
logging.info(f” Hausdorff Distance: {hausdorff:.4f} mm")
logging.info(f" Overlap Fraction: {overlap:.4f}“)
except Exception as e:
logging.error(f"Error computing metrics for {organ}: {e}”)
if name == “main”:
main()