I am trying to register two 3D CT scans using SimpleITK. After checking they have the same orientation, I load them from .nii.gz files. Then they look like this (central axial slice displayed):
Fixed image:
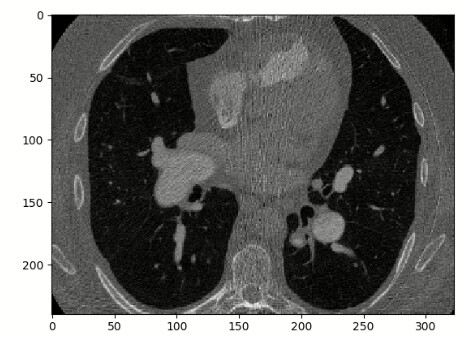
Moving image:
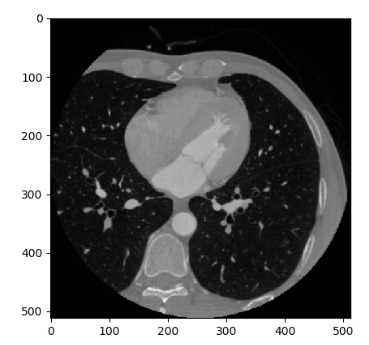
To initialize the registration, following this tutorial we first need to align the two volumes. However, after doing this:
initial_transform = sitk.CenteredTransformInitializer(fixed_image,
moving_image,
sitk.Euler3DTransform(),
sitk.CenteredTransformInitializerFilter.GEOMETRY
)
moving_resampled = sitk.Resample(moving_image, fixed_image, initial_transform, sitk.sitkLinear, 0.0, moving_image.GetPixelID())
The moving_resampled image is flipped upside down:
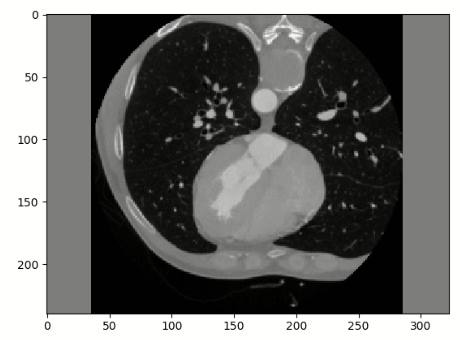
Is this normal behavior?
As an alternative, I tried to just align the images using a 3D translation, which gives an error:
initial_transform = sitk.CenteredTransformInitializer(fixed_image,
moving_image,
sitk.TranslationTransform(3),#sitk.Euler3DTransform(),
sitk.CenteredTransformInitializerFilter.GEOMETRY,
) # .MOMENTS)
moving_resampled = sitk.Resample(moving_image, fixed_image, initial_transform, sitk.sitkLinear, 0.0, moving_image.GetPixelID())
File “C:\Users\stijnb\AppData\Local\Continuum\miniconda3\envs\mimicsenv352\lib\site-packages\SimpleITK\SimpleITK.py”, line 6169, in CenteredTransformInitializer
**return _SimpleITK.CenteredTransformInitializer(*args, kwargs)
RuntimeError: Exception thrown in SimpleITK CenteredTransformInitializer: D:\a\1\sitk\Code\BasicFilters\src\sitkCenteredTransformInitializerFilter.cxx:132:
sitk::ERROR: Error converting input transform to required transform type with center.
Any suggestions how this can be solved? Thanks!