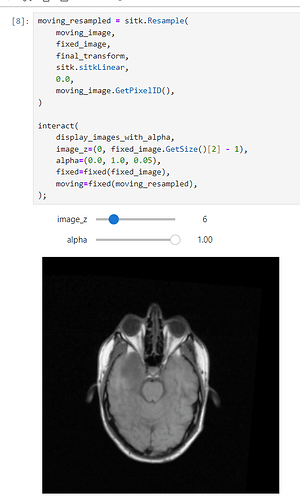
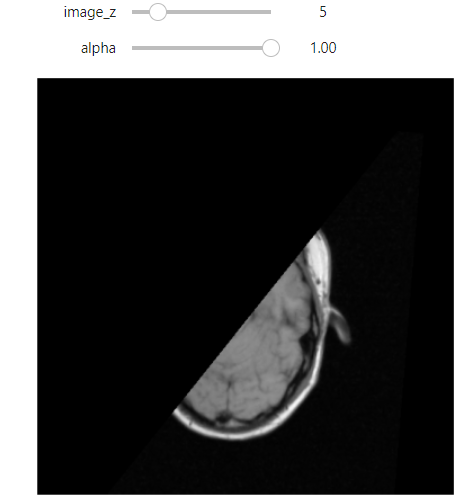
Hi @zivy,
Thank you for your response. I did have a look at the notebook you sent me, and I have already tried sometime similar to calculate the output size of the resampling grid.
Here’s a snippet of my code, that I took from the toolkit’s Transforms and Resampling notebook , and adapted to my 3D images.
import SimpleITK as sitk
import numpy as np
import itertools
def compute_common_bounding_box(images, transforms):
"""
Compute the axis-aligned bounding box that encompasses all transformed images.
Parameters:
images (list): List of SimpleITK image objects.
transforms (list): List of corresponding transforms for each image.
Returns:
tuple: A tuple containing new_origin, new_size, new_spacing, new_pixel_type, and new_direction.
"""
dim = images[0].GetDimension()
boundary_points = []
for image, transform in zip(images, transforms):
# Consider all corners of the image in 3D
for boundary_index in itertools.product(*zip([0] * dim, [sz - 1 for sz in image.GetSize()])):
# Convert index to physical space
physical_point = image.TransformIndexToPhysicalPoint(boundary_index)
# Transform the point to the fixed image coordinate system
transformed_point = transform.GetInverse().TransformPoint(physical_point)
boundary_points.append(transformed_point)
# Calculate the bounding box coordinates
max_coords = np.max(boundary_points, axis=0)
min_coords = np.min(boundary_points, axis=0)
new_origin = min_coords
new_spacing = images[0].GetSpacing()
new_pixel_type = images[0].GetPixelID()
new_size = (((max_coords - min_coords) / new_spacing).round().astype(int)).tolist()
new_direction = np.identity(dim).ravel()
return new_origin, new_size, new_spacing, new_pixel_type, new_direction
def resample_images_to_common_grid(images, transforms):
"""
Resample images onto a common grid.
Parameters:
images (list): List of SimpleITK image objects.
transforms (list): List of corresponding transforms for each image.
Returns:
list: List of resampled SimpleITK image objects.
"""
# Compute bounding box for all images
new_origin, new_size, new_spacing, new_pixel_type, new_direction = compute_common_bounding_box(images, transforms)
resampled_images = []
for image, transform in zip(images, transforms):
resampled_image = sitk.Resample(
image,
new_size,
transform,
sitk.sitkLinear,
new_origin,
new_spacing,
new_direction,
0.0,
new_pixel_type,
)
resampled_images.append(resampled_image)
return resampled_images
# Example usage:
fixed_image = sitk.ReadImage('path_to_fixed_image.nii')
moving_image = sitk.ReadImage('path_to_moving_image.nii')
# Replace with the actual registration result transform
registration_result = sitk.Transform(3, sitk.sitkIdentity)
images = [fixed_image, moving_image]
transforms = [sitk.Transform(3, sitk.sitkIdentity), registration_result]
resampled_images = resample_images_to_common_grid(images, transforms)
I tested this on both the MRI data from 60_Registration_Introduction.ipynb and my own, but some 2d slices in the boudaries of the moving image are still being mapped out of domain (the fixed image as well):
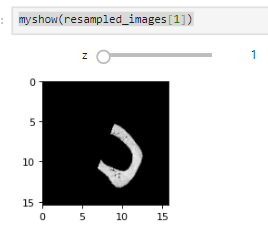
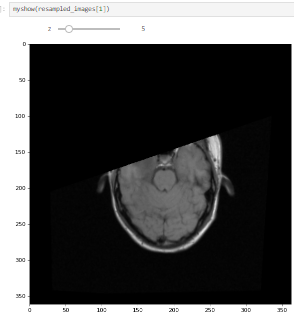
What am I doing wrong? I compute the common bounding box from both images, measure the new resampling grid’s dimension, and resample both images to this new grid.
Thanks a lot for your guidance and efforts.