Hello,
I was performing registration between two modalities of brain and got weird results. It is hard to describe. It looks like some of the edges are highlighted. Is that normal?
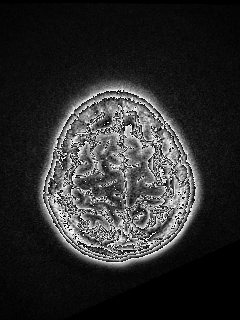
Here is my code.
Hello,
I was performing registration between two modalities of brain and got weird results. It is hard to describe. It looks like some of the edges are highlighted. Is that normal?


Here is my code.
Hello @Jennsoo,
This isn’t a registration problem, it is a visualization/image-storage format issue. The moving_resampled image has a pixel type of sitkFloat32. The original/native type is likely integral (int16 or uint16). You will need to save it appropriately, or use a visualization program that does the correct thing with float data.
See if this helps:
moving_resampled = sitk.Resample(sitk.ReadImage(sitk._mageSeriesReader_GetGDCMSeriesFileNames(t2_path, moving_ids[0]), fixed_image, final_transform, sitk.sitkLinear, 0.0)
What software did you use for visualization? Try software that is specifically designed for medical image visualization, such as 3D Slicer or ITK-snap.
I did not use any software for visualization. I need to save every slice into a single .png/jpg/… So I transfered the moving_image into UInt8 type using sitk.Cast.
Hello,
Thank you for your constructive suggestions. It is exactly the image-storage format issue. I checked the notebook again and found that I forgot a step when storing the results, the sitk.RescaleIntensity function. Forgive my silly mistake!
images = sitk.Cast(sitk.RescaleIntensity(moving_resampled), sitk.sitkUInt8)
Hello @Jennsoo,
Please be careful with this as you are loosing information. Common, non medical imaging formats (png, jpg…) are not really suitable for use with medical images. Save your data in these formats only if you intend to use it for a figure in a manuscript. Otherwise, save in a format that supports high dynamic range (nrrd, mha,…) and use an appropriate viewer as recommended by @lassoan.