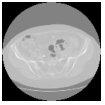
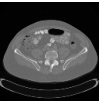
Hello,
I am working on 3D CT images. I need to perform registration on two different datasets. I randomly selected a scan from dataset 1 as the fixed_image and perform a 3d affine registration to register all the scans from dataset 2 to it. My code is as follows:
def register(fixed_image, moving_image):
initial_transform = sitk.CenteredTransformInitializer(fixed_image,
moving_image,
sitk.AffineTransform(fixed_image.GetDimension()),
sitk.CenteredTransformInitializerFilter.GEOMETRY)
moving_resampled = sitk.Resample(moving_image, fixed_image, initial_transform, sitk.sitkLinear, -1000, moving_image.GetPixelID())
registration_method = sitk.ImageRegistrationMethod()
registration_method.SetMetricAsMattesMutualInformation(numberOfHistogramBins=50)
registration_method.SetMetricSamplingStrategy(registration_method.RANDOM)
registration_method.SetMetricSamplingPercentage(0.01)
registration_method.SetInterpolator(sitk.sitkLinear)
registration_method.SetOptimizerAsGradientDescent(learningRate=1.0, numberOfIterations=500, convergenceMinimumValue=1e-6,
convergenceWindowSize=10)
registration_method.SetOptimizerScalesFromPhysicalShift()
registration_method.SetInitialTransform(initial_transform, inPlace=False)
registration_method.AddCommand(sitk.sitkStartEvent, start_plot)
registration_method.AddCommand(sitk.sitkEndEvent, end_plot)
registration_method.AddCommand(sitk.sitkMultiResolutionIterationEvent, update_multires_iterations)
registration_method.AddCommand(sitk.sitkIterationEvent, lambda: plot_values(registration_method))
final_transform = registration_method.Execute(sitk.Cast(fixed_image, sitk.sitkFloat32),
sitk.Cast(moving_image, sitk.sitkFloat32))
moving_resampled = sitk.Resample(moving_image, fixed_image, final_transform, sitk.sitkLinear,-1000,moving_image.GetPixelID())
moving_resampled_npa = sitk.GetArrayFromImage(moving_resampled)
return moving_resampled_npa
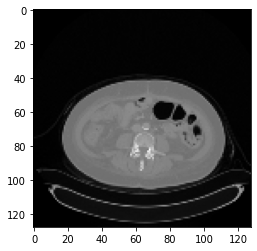
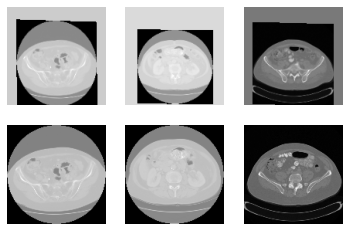
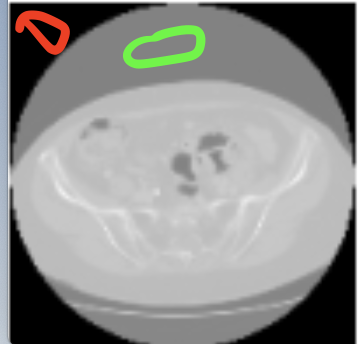
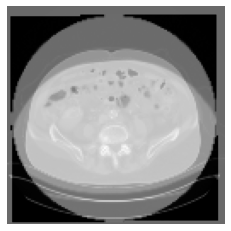
However, after registration there is a gray color border around some images. How can I avoid it?
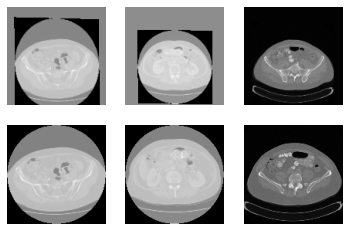
In the above image, first row is after registration. Second row is before registration.